
Andrew Felton, PhD, Vice President of Marketing, Clinical Next Generation Sequencing
The availability of a next-generation sequencing (NGS) assay using cfDNA input is enabling cancer researchers to accelerate their work to help move the community closer to personalized medicine. We recently caught up with Andrew Felton to chat about his views on cell-free DNA (cfDNA) cancer research.
Why are you excited about cfDNA for cancer research?
The idea of detecting the mutations involved in cancer from a simple blood draw, has been an aspirational goal for the oncology community for a considerable period of time. I’m truly excited to see the realization of this dream come to fruition for cancer research, and for our team to contribute to this progress. It is really exciting and rewarding to finally be able offer a solution to the community, and then observe the ingenious work begin to be demonstrated and published by our customers.
To that end, we recently released a new liquid biopsy NGS assay. What’s special about the Oncomine™ Lung cfDNA Assay?
The Oncomine™ Lung cfDNA Assay is a tumor type-specific, multi-biomarker next-generation sequencing (NGS) assay that, from a single tube of blood, enables detection of somatic mutations in plasma, down to a level of 0.1% in genes relevant to solid tumors.
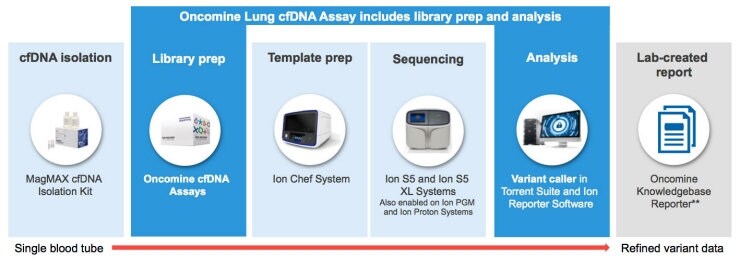
The assay is part of a complete solution for cancer research enabling the detection of cell free DNA (cfDNA) derived from tumor cells that may be freely circulating in the blood. Our product includes the reagents for library construction as well as a single pool of primers used to perform multiplex PCR for preparation of amplicon libraries from cfDNA found in the plasma fraction of whole blood.
The assay targets key SNVs and indels to help researchers analyze and better understand the actions and effects of tumor drivers and resistance mutations, and is uniquely capable at detecting rare variants in cfDNA with very high specificity based on the tag sequencing technology.
You mention the assay enables a limit of detection at 0.1%. How is that significant?
When we talk to our customers about using blood samples in their research, many of them tell us they need an NGS assay, a multi-biomarker assay, capable of detecting variants below 1%. What’s particularly interesting about the Oncomine Lung cfDNA Assay is that it’s flexible; we’ve been able to achieve robust results from 30ng input at 0.05%* all the way down to 1ng input at 0.6% limit of detection.
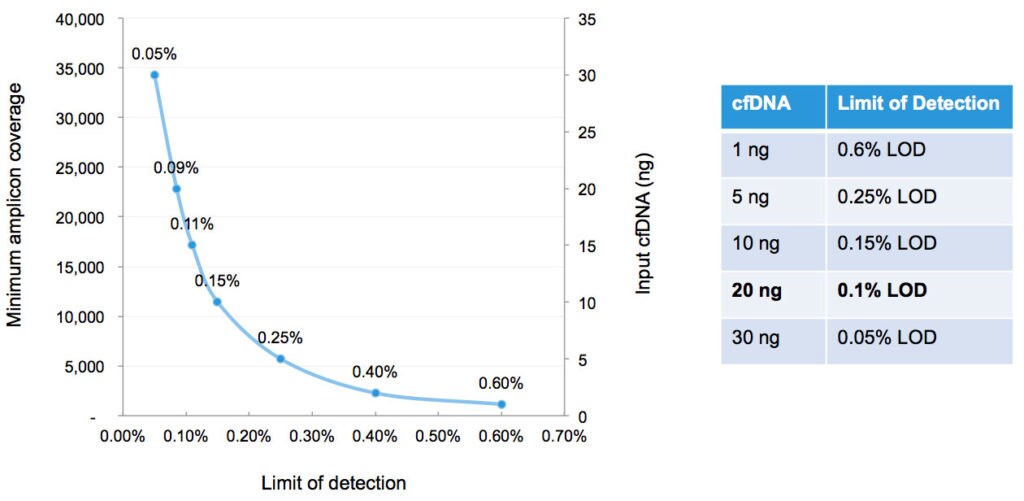 That’s critical because when you are dealing with precious cancer research samples, many researchers don’t have access to upward of 20ng to 30ng of starting material in order to detect the minor variant components that may be present in the sample.. They may only have 5ng of starting material or less. With this new assay, our customers are enabled to get answers from more samples, even from those samples where very little starting material may be available.
That’s critical because when you are dealing with precious cancer research samples, many researchers don’t have access to upward of 20ng to 30ng of starting material in order to detect the minor variant components that may be present in the sample.. They may only have 5ng of starting material or less. With this new assay, our customers are enabled to get answers from more samples, even from those samples where very little starting material may be available.
This assay is enabled by tag sequencing technology. What is it?
Tag sequencing technology builds on the strength of next-generation sequencing to enable researchers to focus on data analysis and interpretation while harnessing sequencing power to look at multiple targets at once.
Using tag sequencing in the Oncomine Lung cfDNA Assay, we’re able to attach unique molecular tags to short DNA sequences prior to amplification. After sequencing, our analysis software then aligns, groups, and filters reads based on families of molecules that originated from the same source cfDNA molecule. This helps researchers detect interesting hotspots and indels and to filter out false positives to achieve highly accurate variant calls down to 0.1% limit of detection.
Tag sequencing technology is designed to address specific biological challenges present in cell-free DNA—cfDNA is highly fragmented and the circulating tumor DNA component is very rare. The technology leverages the same Ion Torrent sequencers, such as the Ion S5™ Series Systems, and Ion Chef™ System, and workflows our customers have used for years.
How do you see this assay being used in the near term?
Translational cancer researchers are starting to use research blood samples to better understand tumor drivers, resistance mutations, and recurrence. This assay helps researchers assess the viability, reliability, and implications of using a blood sample both in concert with, and in lieu of tissue samples.
We’ve designed the cfDNA lung assay to have complementary content to our solid-tissue assays like the Oncomine Focus™ Assay or the Ion AmpliSeq™ Cancer Hotspot Panel v2, with consistent gene coverage to enable concordance studies, longitudinal assessment of recurrence and ongoing analyses.
For example, from library prep, to automated templating using the Ion Chef System, to NGS on the Ion S5 System, to data analysis using Ion Reporter™ software, the Oncomine™ oncology workflow helps our customers detect low-frequency mutations in research samples, to better follow resistance as it emerges. And, we also offer the Oncomine™ Knowledgebase Reporter, a specialty software report for the assay which helps customers narrow down relevant information from hundreds, down to a few relevant cancer driver mutations.
Isn’t “Oncomine” the assay used in the NCI-MATCH trial?
Oncomine isn’t one assay; Oncomine branded products are part of a suite designed specifically for oncology. It is the Oncomine Comprehensive Assay, which is based on Ion AmpliSeq™ technology, that is being utilized to test and enroll patients in the NCI-MATCH trial—a nationwide US trial to analyze 3,000 patient samples across multiple cancer types. The trial has recently moved into its 2nd phase, which increases the number of enrolled patients to 5,000 and adds several new treatment arms to the study.
The Oncomine brand and products represent our premium oncology product line; designed to help accelerate cancer clinical research, they offer solutions with protocols based on verification using clinical samples, and content consisting of key cancer driver genes and optimized bioinformatics tools including reporting of relevant variants.
Many other companies have cfDNA services. Why did you develop a cfDNA assay and not a service?
Outsourcing samples may be a good option for some customers. We see a number of advantages for customers if they choose to analyze their samples in their own labs. The Oncomine Lung cfDNA Assay workflow is simple and fast; and we’ve designed the assay to be accurate, to help researchers regain control of their blood samples without sacrificing performance. NGS analysis of plasma samples at 0.1% limit of detection was a need among pathologists looking at liquid biopsy techniques that could enable more timely information about a tumor; information that in the future, may help change treatment decisions.
For More Information:
Oncomine Lung cfDNA Assay Workflow click here
Download Oncomine Lung cfDNA Assay Workflow here
Ion Ampliseq Technology click here
Liquid Biopsy Cancer Research click here
Next-Generation Sequencing Platforms click here
* The Oncomine Lung cfDNA Assay is verified at 0.1% limit of detection with 20 ng input DNA.
For Research Use Only. Not for use in diagnostic procedures.
Well that will be a new chapter in medical diagnostics….