What is multiplexing?
Multiplexing, in biological applications, is defined by the simultaneous evaluation of several experimental elements, thereby increasing the throughput of analysis and reducing the burden of time and cost associated with the investigation of those individual components. [1-3] Increasingly, researchers are combining various assays to interrogate complex biological questions related to gene expression profiles, signaling pathway events, and cell health and proliferation indicators, to name a few. [4-8] Through advanced technologies, using flow cytometry, imaging, plate reader, and mass spectrometry formats, multiparameter analysis of a given sample, at the single cell level, is made possible. [6-11] Further coupling of technologies, as with RNA detection through branched DNA amplification and immunolabeling, or with acoustic cytometry and distinct fluorescence emitting probes, will continue to expand the number of parameters that can be analyzed concomitantly. This review highlights a few multiplexed applications developed by Thermo Fisher Scientific, to help biomedical researchers overcome many of the limitations caused by sample scarcity, or variability across samples and platforms. Navigate to the related BioProbes articles for a more detailed discussion around each topic area.
Simultaneous visualization of protein and RNA expression at the single-cell level
Invitrogen PrimeFlow RNA and ViewRNA Cell Plus are two assays that effectively combine various technologies to study RNA molecules on a single-cell level. Compatible with the Cellnsight CX7 High-Content Screening platform, the ViewRNA Cell Plus assay combines ViewRNA ISH technology, a proprietary fluorescent in situ hybridization (FISH) and branched DNA (bDNA) amplification technique, with antibody-based protein detection to simultaneously visualize RNA and protein in single cells. ViewRNA Cell Plus assay made traditional incompatible immunocytochemistry (ICC) and in situ hybridization (ISH) compatible. Similarly, the PrimeFlow RNA assay employs FISH with bDNA signal amplification for the simultaneous detection of up to four RNA targets. This can be used in combination with immunolabeling for both cell-surface and intracellular proteins detection using fluorophore-conjugated antibodies for analysis by flow cytometry.

Figure 1. The ViewRNA Cell Plus Assay workflow.The workflow for the Invitrogen ViewRNA Cell Plus Assay Kit starts with fixation, permeabilization, and antibody labeling, followed by hybridization with RNA-specific target probes. This hybridization is then detected after branched DNA (bDNA) signal amplification using preamplifiers, amplifiers, and label probes. Labeled cells are analyzed on a fluorescence microscope or high-content imager.

Figure 2. The PrimeFlow RNA assay workflow.The workflow for the Invitrogen PrimeFlow RNA Assay Kit starts with optional antibody labeling, followed by fixation and permeabilization, and then hybridization with gene-specific target probes. This hybridization is then detected after branched DNA (bDNA) signal amplification using preamplifiers, amplifiers, and label probes.
Interested in learning more? Read these BioProbes articles!
BioProbes 75: Evaluate both RNA and protein targets in single cells
BioProbes 76: Double vision: Simultaneous visualization of protein and RNA targets
Robust cell health analysis using multiplexable cell viability assays
Viability assessment is critical in many application areas in basic, pre-clinical and clinical research settings. Viability probes can be multiplexed with a combination of different measurements to provide more cellular information than any single-parameter assay. For instance, Figure 3 demonstrates a workflow to investigate both proliferation and viability by using CyQUANT Direct cell proliferation assay and PrestoBlue viability reagent. CyQUANT direct is a highly sensitive DNA-content based assay and PrestoBlue cell viability reagent is a metabolism-based assay. The use of the PrestoBlue and CyQUANT Direct assays together allow for cell metabolism, DNA content, and changes in membrane permeability, to be assessed in the same sample, using distinct fluorescent signals. The tandem CyQUANT Direct and PrestoBlue protocol is easy to carry out and requires no wash or cell lysis steps.

To perform the multiplex viability assay, add PrestoBlue™ reagent to drug-treated cells, incubate 10 min, and read fluorescence. Following the PrestoBlue readout, add CyQUANT™ Direct reagent to cells, incubate an additional 60 min, and re-read fluorescence.
Discover how! BioProbes 73: CyQUANT direct and PrestoBlue viability assays work together
Explore ways to cooperatively investigate mitochondrial morphology and function
Monitoring changes in mitochondrial morphology and function are valuable indicators of cell health. Mitochondrial morphology reagents, that allow for the assessment of membrane integrity, can be combined with functional probes to provide more information about mitochondrial health. Invitrogen reagents can be used to study mitochondrial membrane potential, calcium flux, oxidative phosphorylation, autophagy/mitophagy and cytosolic pH. Furthermore, improvements in the development of probes has enabled researchers with more fluorescence options that can be combined to study mitochondria. For example, in Figure 4 below, Invitrogen CellLight Mitochondria-GFP or CellLight Mitochondria-RFP can be combined with dyes such as Tetramethylrhodamine, Methyl Ester, Perchlorate (TMRM) to monitor mitochondrial structural integrity while also assessing membrane potential.

HeLa cells were transduced with CellLight™ Mitochondria-GFP and loaded with 50 nM TMRM for 10 min at 37°C. (A–E) Images were acquired at 5 sec intervals for 90 sec following treatment with the uncoupler CCCP; zoomed sections (B–D) reveal heterogeneity in mitochondrial membrane potential regulation. Transient depolarization was observed in one but not all mitochondria (C, arrow), as indicated by loss of orange TMRM signal; GFP fluorescence was maintained during depolarization, indicating an intact mitochondrion. Loss of mitochondrial membrane potential was evident by 90 sec post-CCCP treatment (E); however, mitochondria were still intact, information that would have been lost using TMRM alone.
Read more about mitochondrial health!
BioProbes 72: Tools to study mitochondrial morphology and function
Multiplexable assays for in situ apoptosis detection in combination with multiple fluorescent dyes
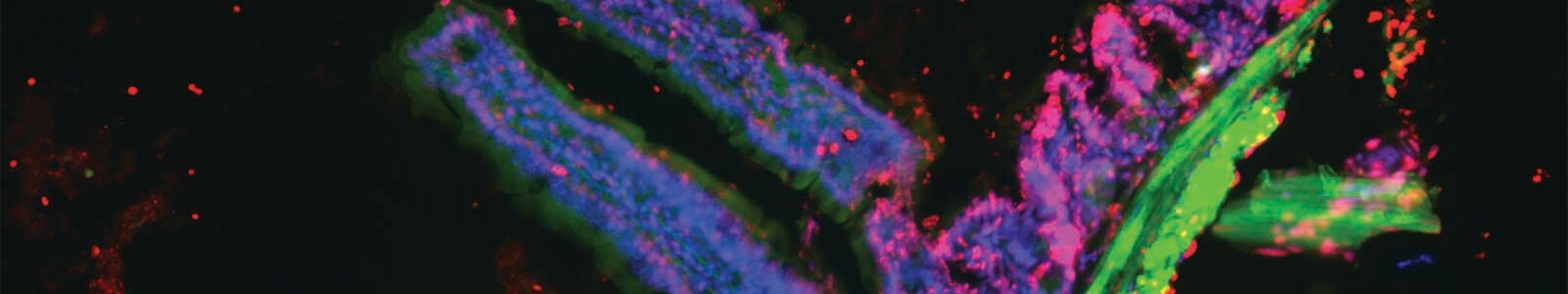
Changes in nuclear morphology, chromatin condensation, nuclear envelope degradation, and fragmentation of cellular DNA are all indicators of late stage apoptosis. Detected is made possible using DNA-binding dyes and in situ hybridization. Invitrogen Click-iT Plus, with terminal deoxynucleotidyl transferase-dUTP nick end labeling (TUNEL) assays, enables researchers to achieve specific and sensitive DNA fragmentation detection that can be multiplexed with other fluorescence-based approaches. The Click-iT Plus TUNEL assay offers gentle reaction and highly sensitive detection conditions that can be applied to wide range of cell types, while including various fluorescent proteins or reagents. Figure 5 below demonstrates the combinatorial of Click-iTTM Plus TUNEL Assay with Alexa FluorTM 594 dye, muscularis externa expressing GFP, nuclei labeled with Thermo Scientific Hoechst 33342 dye, and actin staining with Invitrogen Alexa FluorTM 647 phalloidin in a single tissue section analyzed by microscopy.

Formalin-fixed, paraffin-embedded (FFPE) tissue from a transgenic mouse expressing GFP in intestinal muscle was treated with DNase I. (A) Cell nuclei are stained by Hoechst 33342 dye (blue), (B) the GFP signal (green) is detected in the surrounding muscular layer, (C) filamentous actin is stained by Alexa Fluor 647 Phalloidin (purple), and (D) the TUNEL-positive signal resulting from DNase I treatment is clearly defined by the Click-iT Plus TUNEL Assay with Alexa Fluor 594 dye (red). The last panel (E) is the multiplexed image resulting from an overlay of the four fluorescent signals.
Find out more!
BioProbes 72: Multiplexable Click-iT Plus TUNEL assays for in situ apoptosis
___________________________________________________________________________________________________________________________________________________________________________________________
References
- Krutzik PO, et.al., Fluorescent cell barcoding for multiplex flow cytometry. Curr Protoc Cytom 2011; Chapter:Unit6.31.
- Bradford JA, et.al., Fluorescence-intensity multiplexing: Simultaneous seven-marker, two-color immunophenotyping using flow cytometry. Cytometry A 2004; 61A:142–152.
- Smurthwaite C, et.al. Fluorescent genetic barcoding in mammalian cells for enhanced multiplexing capabilities in flow cytometry. Cytometry A. 2013; 85A:105-113.
- Grant GD, et.al., Live-cell monitoring of periodic gene expression in synchronous human cells identifies Forkhead genes involved in cell cycle control. Mol Biol Cell 2012; 23:3079–3093.
- Lu R, et.al., Tracking single hematopoietic stem cells in-vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol 2011; 29:928–933.
- Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods 2006; 3:361–368.
- Edwards BS, et.al.,. High-throughput cytotoxicity screening by propidium iodide staining. Curr Protoc Cytom 2007; Chapter 9:Unit9.24.
- Violin JD, et.al., A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol 2003; 161:899–909.
- Ashcroft RG, Lopez PA. Commercial high-speed machines open new opportunities in high throughput flow cytometry (HTFC). J Immunol Methods 2000; 243:13–24.
- Edwards BS, et.al., High-throughput flow cytometry for drug discovery. Expert Opin Drug Discov 2007; 2:685–696
- Barteneva NS, et.al., Imaging flow cytometry. Coping with heterogeneity in biological systems. J Histochem Cytochem. 20012; Chapter 60:Unit10:723-733.
I don’t ordinarily comment but I gotta state thanks for the post
on this special one :D.