 Biobanking has an important role to play in discovering the molecular basis of disease. Researchers need access to representative libraries of biospecimens, and these samples need to be preserved in excellent quality for valid genomic investigation. Lee et al. (2015) took a closer look at biobanking ileum mucosa samples for genomic studies, paying close attention to conditions surrounding tissue collection that might adversely affect gene and RNA stability.1
Biobanking has an important role to play in discovering the molecular basis of disease. Researchers need access to representative libraries of biospecimens, and these samples need to be preserved in excellent quality for valid genomic investigation. Lee et al. (2015) took a closer look at biobanking ileum mucosa samples for genomic studies, paying close attention to conditions surrounding tissue collection that might adversely affect gene and RNA stability.1
In their study, Lee et al. examined the impact of warm ischemia (at body temperature or 37°C) or cold ischemia (placed on ice on removal from the donor’s body) on RNA quality during storage in an RNA stabilizing solution or in snap-frozen samples. Noting that studies into RNA stability in ileum mucosa tissue were not available, the team worked with surgical teams treating patients with colon carcinoma, where normal intestinal tissue was available during the standard hemicolectomy procedure.
Working alongside surgeons in the operating room, biobanking staff took control of the normal ileum sections once removed from the patient. In this way, the researchers could control for the type of (warm or cold) ischemia and the duration of tissue exposure prior to placement in storage for RNA studies (snap frozen versus submersion in RNAlater solution). From baseline studies carried out over 52 collections, Lee et al. determined that normal processing of ileum mucosa tissue by the surgical team resulted in exposure to warm ischemia for 12±13 minutes and to cold ischemia for 22±15 minutes. They therefore examined tissue collected from six donors at the following time points:
- Warm ischemia = 0, 15, 30, 60, 90 minutes prior to snap freezing or placing in RNAlater solution
- Cold ischemia = 0, 30, 60, 90, 180, 360 minutes prior to snap freezing or placing in RNAlater solution
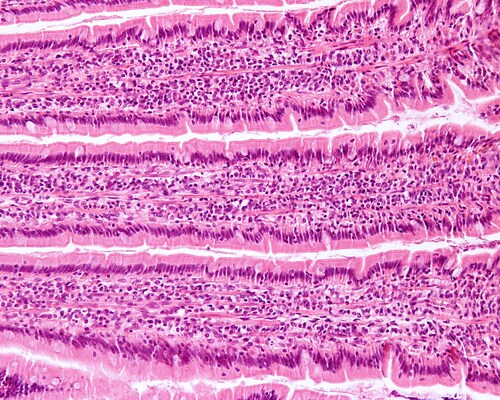
The researchers also prepared formalin-fixed, paraffin-embedded sections for subsequent hematoxylin and eosin staining to examine cell architecture and integrity for each time point. They found good tissue morphology at each time point and storage medium, with intact ileum epithelium retained for all samples.
The research team examined nucleic acid purity using a NanoDrop 2000 spectrophotometer (Thermo Scientific). They found that purity was good enough for onward genomic investigation. Once the team established sample quality, they extracted the RNA using a commercial kit, then measured its integrity by chip analysis to generate an RNA integrity number (RIN). RIN values did not differ significantly across time points or storage mediums, although samples stored in the RNAlater solution showed higher values.
Next, the research team assessed RNA stability by examining a set of reference genes using real time-quantitative polymerase chain reaction (RT-PCR). They used the StepOnePlus Real-Time PCR System along with TaqMan Gene Expression Assays (both Thermo Scientific) to examine HPRT1, GUSB, PSMB6, TBP and RPL13 as reference genes, and then four inducible genes, MYC, HIF1α, IL1β and HMOX1. Pairwise analysis of the reference genes showed no significant differences between samples and time points, although this analysis did identify RP13 as the least stable. Lee et al. did not find any significant differences in levels of the four inducible genes with either ischemia or storage method.
In conclusion, Lee et al. suggest that the findings show that gene expression in ileum mucosa samples is not affected by warm or cold ischemia over the time points studied. Furthermore, they saw no difference in storing the samples by snap freezing or in an RNA stabilizing solution, suggesting only that biobankers consider onward applications when choosing biobanking protocols.
Reference
1. Lee, S.M.L, et al. (2015) “Pre-analytical determination of the effect of extended warm or cold ischaemia on RNA stability in the human ileum mucosa,” Public Library of Science PLoS ONE 10(9): e0138214. doi:10.1371/journal.pone.0138214
Leave a Reply