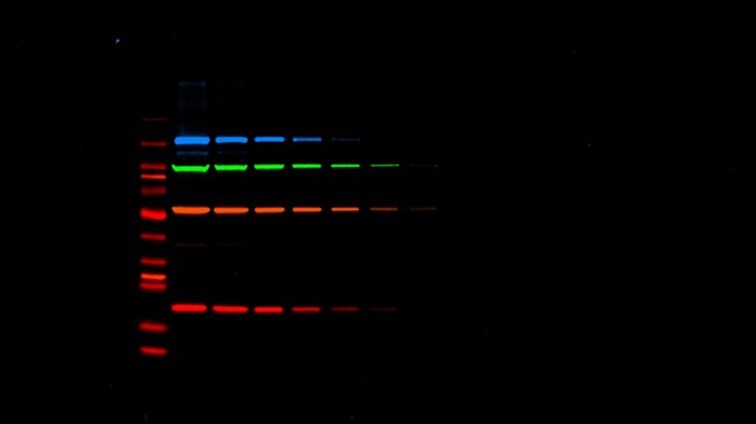
Fluorescence detection multiplexing achieves more data from your sample with iBright. Multiplexing can capture up to four proteins in a single blot for more meaningful and representative experiments, a must for western blot publication.
For today’s top journals, total protein normalization and high-quality images are the keys to western blot publication
Table of Contents
Introduction
Overview of Normalization
Housekeeping Proteins vs. Total Protein Normalization
Journal-Specific Western Blot Publication Guidelines
Nature
Science
Cell Press
Journal of Biological Chemistry
Elsevier
Wiley
MDPI
Journal of Cell Biology
American Association of Cancer Research (AACR)
Common Mistakes in Western Blot Figures
More Resources for Western Blot Reporting
Western blotting is a powerful tool in the sense that it can tease out the needle – a single protein of interest – in a proverbial haystack sample containing hundreds of proteins.
For more than 40 years, the western blot or immunoblot has been a workhorse of the wet lab. In its early days, the assay delivered information in a strict binary of ‘yes’ or ‘no’ – protein present or absent.
In recent years, however, improved western blot technology and technique have allowed an exciting quantitative element to emerge. Researchers can now reliably measure relative changes in protein expression and abundance.
But with this shift towards quantitative understanding comes a movement by major journals, funding agencies, and professional scientific societies to uphold the highest standards of data integrity for publication. Their updated guidelines for immunoblotting images come alongside guardrails against data manipulation, intentional or inadvertent, in a growing age of generative AI and accessible image editing software.
So, what does a “publication-ready” western blot mean today to the top journals in the field, and what tips, tools, and products exist to help researchers get there? This comprehensive guide will help steer you through the process.
Quantitative Western Blotting Requires Normalization
With a quantitative western blot, researchers can gain valuable insight into relative protein expression.
Quantitative western blotting requires careful interpretation, particularly around non-uniformities. Variability in western blotting occurs most often with unequal protein concentrations, inconsistent sample loading onto the gel, and irregularities during transfer – all normal, human realities of the process.
Normalization accounts for those realities by distinguishing experimental variability from true biological changes in protein expression. Skillful normalization ultimately encourages accuracy and reproducibility in the literature.
Right now, methods for immunoblotting normalization and quantitation across the field vary more than many experts would like.
Editorial staff of the field-leading publication Journal of Biological Chemistry named acceptable presentation and quantitation of western blots as one of three “major gaps in overall data reporting” among submissions. Their recently issued revised guidelines for blot reporting reflect a wider push towards total protein normalization (TPN) in lieu of another popular normalization strategy using housekeeping proteins (HKP).
Let’s talk through some of the downsides of housekeeping protein protocols and advantages of total protein normalization.
Housekeeping Proteins (HKP) Are Falling Out of Favor with Journals
For decades, researchers have used housekeeping proteins like GAPDH, β-tubulin, β-actin, or cyclophilin B as internal loading controls for normalization. In this approach, changes in a sample’s target protein level can be expressed as a ratio to the chosen HKP.
HKP is still the most-used method of western blotting normalization today.
The problem with this protocol is that it assumes consistent HKP expression across experiments, and more and more studies have now confirmed that this just isn’t the case. HKP expression is more variable than constant, changing with cell type and developmental stage; tissue age, type, and pathology; post-transcriptional regulation; experimental conditions; and more (Table 1). HKPs are also typically much more abundant than target proteins, meaning that band intensities can saturate easily and early in the HKP control and lead to misinterpretations. In Figure 1 below, notice that signal saturation begins at 30 µg for loads with common HKP’s GAPDH, β-actin, and α-Tubulin.

Figure 1. Total protein normalization using the No-Stain Protein Labeling Reagent: Bolt 4-12% Bis-Tris Plus gels were loaded with HeLa Lysate ranging from 10 to 50 µg and electrophoresed using MES running buffer. Proteins from the gels were transferred onto PVDF membranes using the Invitrogen iBlot 2 Gel Transfer Device with iBlot 2 Transfer Stacks, PVDF, mini (P0 protocol for 7 minutes). The PVDF membranes were washed twice for 2 minutes with 20 mL of ultra-pure water on a rotating platform, whereupon they were labeled with 10 mL of No-Stain labeling solution on a rotating platform for 10 minutes. The membranes were then washed 3 times for 2 minutes with 20 mL of ultra-pure water on a rotating platform, followed by immunoblotting for β-actin (Cat. no. AM4302), GAPDH (Cat. no. 398600), and α-tubulin (Cat. no. 138000) followed by goat anti-mouse Alexa Fluor Plus 680 (Cat. no. A21058). The blot was imaged using the iBright Imager. The iBright software was used to quantitate the total protein signal in the lanes. The linear regression value of the plotted data for the entire load range using the No-Stain Protein Labeling Reagent was determined (R2 = 0.9990), whereas the R2 values for β-actin, GAPDH, and α-tubulin were 0.8851, 0.9438, and 0.8332, respectively.
HKP normalization is also a lot of work. It requires laborious setup of positive and negative controls, along with optimizing for a linear dynamic range of detection in which band signal intensity of both HKP and target protein are directly proportional to the amount of protein loaded. This linear range can be prohibitively narrow or non-existent.
HKPs can also interact with experimental components in problematic ways – co-migrating with similar-sized target proteins, for example, or requiring antibodies that cross-react with target proteins.
Total Protein Normalization (TPN) is the New Gold Standard for Western Blot Quantitation

Figure 2. Quantitative analysis using the No-Stain Protein Labeling Reagent and an iBright Imaging System. A Novex 4-12% Tris-Glycine gel, WedgeWell format, was loaded and electrophoresed with lysates from HeLa cells expressing RB1, at total protein loads ranging from 0.6 to 10 µg. Proteins from the gel were transferred to a nitrocellulose membrane using the iBlot 2 Dry Blotting System. The nitrocellulose membrane was labeled using the No-Stain Protein Labeling Reagent for 10 minutes and the labeled membrane was imaged using the iBright imager. The same No-Stain labeled membrane was used to probe for RB1 with a specific antibody labeled with Alexa Fluor 645 dye. The iBright normalization software was used to quantify the total protein signal in lanes loaded with HeLa lysate loads ranging from 0.6 to 10 µg and signal intensities from RB1 immunodetection bands. The signal intensity from the total protein load and RB1 were plotted.
Luckily, HKP is not the only technique for normalization. Total protein normalization (TPN) normalizes the quantity of target protein to the total amount of protein in each lane rather than a single loading control.
TPN is not affected by experimental manipulations, provides a larger dynamic range for detection, and can provide information about the quality of your electrophoresis and blotting setups. For these reasons, it is the superior method for accurate quantitation and increasingly being required by publications.
You can achieve TPN with either a total protein stain or labeling technology. The total protein labeling route is especially compelling because it is fast, easy to perform, and highly sensitive. Compared to total protein stains, a fluorogenic labeling method allows for visualization of a strong, uniform signal with low background and no de-staining steps.
R&D scientists at Thermo Fisher Scientific designed the Invitrogen™ No-Stain™ Protein Labeling Reagent for streamlined, rapid fluorescent labelling of total protein within a gel or on a western blot membrane, followed ideally by high-resolution imaging on an iBright Imaging System.
See how total protein normalization (TPN) is done on an iBright Imaging System with No-Stain Protein Labeling Reagent
» Read Our Protocol: “Biological validation of a novel process and product for quantitating western blots” in Journal of Biotechnology
Journal-Specific Western Blot Publication Guidelines
While publications vary on preferred file format, sizing, and naming conventions, the top journals in biology want transparency and quality in your data.
If you want to leave your options open and be prepared for the strictest submission standards from the get-go, opt for total protein normalization and keep careful track of all experimental parameters and original, untouched images.
The information below summarizes some of the specific figure requirements from popular journals in immunology and protein biology:
Nature
- Figure File Specifications: Nature’s author guidelines for initial submission request RGB color images in 300 dpi or higher resolution, while avoiding very large, print-quality files. Authors can present figures in-line with text in a single Word .docx or PDF file.
- Figure Style: See Nature’s Initial Submission Guidelines and ‘Figures’ section of Formatting Guide for comprehensive information on spacing, font, style, et. al. Figures should include legends of <300 words each, with error bars and statistics defined within.
- Blot Guidelines: See full list of guidance on Nature. Guidelines strongly discourage quantitative comparisons between samples on different gels/blots and require that re-arranged lanes be clearly indicated and cropped gels retain all important bands. Loading controls (e.g. GADPH, actin) must be run on the same blot. High-contrast gels and blots are discouraged, as extended exposure times may mask additional protein bands. Authors should take care to check manuscripts for figure duplications, lane splicing, correct labeling or sample or loading controls, correct matching of unprocessed scans with figures. The ISME Journal, published by Nature Publishing Group on behalf of ISME, has separate editorial policies including guidance on quantitative blots.
- Imaging Editing Policies: The use of touch-up tools such as cloning and healing tools in Photoshop, or any feature that deliberately obscures manipulations, is unacceptable per Nature policy See the Policy on Image Integrity for full guidelines.
- Data Reporting Requirements: See Nature’s Editorial Policies page for information on data reporting requirements.
Science
- Figure File Specifications: Science prefers a single .docx Word file that includes all figures and tables with legends and encourages use of their Word template. PowerPoint files are not accepted. Images should be 300 dpi and CMYK at initial submission stage.
- Figure Style: See Science Author Portal for figure preparation instructions including information on spacing, font, style, et. al. Figure legends should be no longer than 200 words.
- Blot Guidelines: No specific blot guidelines outlined. See Research Standards in Editorial Policy for more information universal data quality policies.
- Image Editing Policies: Science does not allow certain electronic enhancements or manipulations of gels. See “Modifications of figures.” Science editors may use Proofig to screen images for AI manipulation.
Cell Press
- Figure File Specifications: Figures should be submitted as separate files rather than embedded in your written manuscript. For initial submission, Cell Press prefers TIFF or PDF (<3 MB) files, but will also accept JPEG or EPS. Upload as 300 dpi RGB-encoded color images.
- Figure Style: See Cell Press Digital Image Guidelines.
- Image Editing Policies: Keep image processing minimal and ensure that all processing is transparent and explained in figure legends. If you remove lanes from gels and blots or consolidate your data in any way, you must make the alterations obvious. All accepted papers are screened for image irregularities. See all Information for Authors.
Journal of Biological Chemistry
- Figure File Specifications: Figures should be submitted as separate files rather than embedded in your written manuscript. It is the authors’ responsibility to verify the quality of the graphics and confirm that compression of the files during the submission process does not distort the images. Upload as 300 dpi RGB-encoded color images. See JBC Digital Art Guidelines for more information.
- Figure Style: Legends should appear within the manuscript file after the References and not within the figure file. Use a consistent and standard font for all figures, like Arial, Helvetica, Symbol, Mathematical Pi, and European Pi.
- Blot Guidelines: JBC released specific guidelines on quantitation and presentation of western blots in 2015, including requirements for description of antibody products and other methods, splicing, overcropping, and the required inclusion of at least one molecular weight marker in electrophoretic gels.
- Image Editing Policies: JBC has adopted and modified the policy of the Journal of Cell Biology, as explained here.
Elsevier
Elsevier provides umbrella author guidelines for artwork submission, but specific journals within may have additional instructions.
- Figure File Specifications: Elsevier recommends EPS, PDF, TIFF, or JPEG file formats, ensuring numbering, type and format is reflected in the file name (e.g. FIG1.TIF = figure 1 in TIFF format). Images should be RGB and 300-500 dpi. See FAQs.
- Figure Style: Figure text should use one of the following standard fonts: Arial, Helvetica, Courier, Symbol, Times, Times New Roman.
- Image Editing Policies: Elsevier’s policy is that “no specific feature within an image may be enhanced, obscured, moved, removed, or introduced. Adjustments of brightness, contrast, or color balance are acceptable, if, and as long as, they do not obscure or eliminate any information present in the original. Manipulating images for improved clarity is acceptable, but manipulation for other purposes could be seen as scientific ethical abuse and will be dealt with accordingly.”
Wiley
Each Wiley journal may provide specific instructions for submissions. Review the journal’s Author Guidelines to ensure that you’ve met all submission requirements. See general Wiley Press guidelines for more information.
- Figure File Specifications: Preferred image file types for images are TIFF, PNG, and EPS, though GIF, JPEG, DOC, PDF, PPT, PSD, PPT, AI, and PS are also acceptable. Each figure should be 300 dpi resolution with a file size less than 10 MB, and all zipped figures should be less than 500 MB. File names should include only the word ‘figure’ and the number (e.g. Figure_1.tiff).
- Figure Style: Wiley recommends a separate figure legend section after references, though legends can be included anywhere as long as they clearly indicate which figure each explains.
- Blot Guidelines: Some Wiley journals will require authors of papers with gel electrophoresis results and/or blots to provide original unprocessed images as Supporting Information for every result for the journal’s files.
- Image Editing Policies: See ‘Ethical Considerations’ section of Figure Preparation Guidelines page.
MDPI
MDPI lists submission guidelines by journal. Many encourage use of a downloadable Microsoft Word file template for submission, which you can access under the journal’s “Instructions for Authors” page.
- Figure File Specifications: MDPI journals like Biology, Cells, and Cancers prefer figures to be placed in-line into the provided Microsoft Word template. Files for figures should also be provided in a single .zip file, preferably as RBG color JPEG, TIFF, EPS, or PDF files of 300 dpi resolution or higher or minimum 1000 pixel width/height.
- Figure Style: All figures should include a short explanatory title and caption. Insert them into the main text close to their first citation.
- Blot Guidelines: Many MDPI journals require Supporting Information inclusion of all original, uncropped, unadjusted blot images at the time of initial submission. See “Original Images for Blots and Gels Requirements” section of Author Instructions for complete information.
Journal of Cell Biology
- Figure File Specifications: Blot Source Data Figures should be provided as individual PDF files (one file per figure) with minimum resolution of 300-600 dpi. Acceptable figure file formats are TIFF, EPS, AI, PSD. File names should be alphanumeric with no spaces or special characters.
- Figure Style: Use a standard font within figures (Arial, Times New Roman, Courier New, or Symbol) in 8 pt or 5 pt for subscript/superscript.
- Blot Guidelines: JCB requires authors to provide Source Data used to generate figures containing gels and western blots, which consists of fully uncropped and unprocessed images. See “Source Data” section of Submission Guidelines for more information. Describe all antibodies in full in the Materials and Methods section, including accession numbers and/or full epitopes or sequences.
- Image Editing Policies: All accepted figures will go through an image screening process prior to publication, as described in the JCB Data integrity and plagiarism section.
American Association for Cancer Research (AACR)
- Figure File Specifications: For initial submission, AACR journals do not impose strict article size or formatting requirements. Authors are encouraged to format their manuscript in a manner that makes it easy for peer reviewers to read and access. Image files should be RGB in EPS, TIFF, AI, PSD, PNG, PS, or JPEG format. PowerPoint files are discouraged but may be acceptable.
- Figure Style: Each figure should be presented in sequential order and adjacent to its legend, ideally where it is first called out in the text. Each figure must have a legend consisting of a short title followed by a description of the data; it must include the number of technical and biological replicates performed, and stain names as applicable.
- Blot Guidelines: AACR Editorial Policies outline specific guidelines for gels and blots.
- Image Editing Policies: AACR is a member of the Committee on Publication Ethics (COPE) and ascribes to the Principles of Transparency and Best Practice in Scholarly Publishing and images are expected to be compliant with our guidelines as well as those of COPE. Detection of images that do not adhere to these guidelines could impact the manuscript’s acceptance and may result in production delays. Be prepared to respond to requests for source data within 7 days.
Avoid These Common Mistakes When Presenting Your Western Blot Images
A 2022 PLoS-published study examined 551 articles containing western blot images published in the top 25% of journals in neuroscience and cell biology. 90% of these papers included only cropped blot images, often with no supplemental blot source data made available to the reader. Methods sections frequently lacked information on the amount of protein loaded on the gel, blocking steps, and antibody labeling protocol, and primary antibody identifiers were often omitted.
Want to buck the trend of incomplete western data? Here are some tips to keep in mind for avoiding the most common mistakes:
- Avoid cosmetic touch-ups. Specks and smudges are a normal part of immunoblotting, and that’s okay! What is typically not okay is digitally altering an image to remove these imperfections from view or to hide extraneous gel bands. Leave your images as is to avoid inadvertent data manipulation. Tools like the iBright Imaging System can help you create high-definition, crisp gel images from the get-go.
- Check your manuscript for placeholders and duplicate figures. We all do it – copy and paste a ‘placeholder’ image and then forget to come back to it later before submission. A final review, particularly by a fresh set of eyes, before manuscript submission can go a long way.
- Save your original data and consider providing it as public supplement to your readers. Transparency is key for trust and reproducibility, and journals will often request raw data files to this end. Make sure to save and organize your original files and have them handy through the submission process.
- Don’t mislead. Often, misleading figures are not intentionally created that way. When in doubt, think: what would fresh or quickly-scanning eyes see here, and is there room for misinterpretation?
- Be careful with splicing and cropping. If splicing or cropping of gels (e.g. to remove an irrelevant lane) is necessary, you should clearly mark it in the image and disclose again in the figure legend. Some publications prohibit splicing different gels together. We’ve noted some of these journals in our table above.
- Never archive blot data in PowerPoint. PowerPoint is a terrible place to archive or store important blot data! The software compresses image files to a lower quality than is often useful to publications, and some journals like Science do not accept PowerPoint files under any circumstances. Keep and store your original TIFF or native format image files for posterity.
- Document all methods, including loading amounts and antibody reporting. When it comes to reproducibility in western blots, the smallest details matter. This is particularly true with primary antibody reporting and protein sample loading amounts. Kroon et. al provide a helpful template for antibody reporting, along with a toolbox to help researchers improve the reproducibility of their data.
- Include critical information in the legend. The figure legend is the place to be transparent about any image editing, splicing, or cropping for the reader.
- Use loading controls properly. According to Cell Press, “More questions get raised about duplication or misuse of loading controls in figures with western blot or RT-PCR data than almost any other issue.” It may go without saying but be sure to run your loading controls in the same gel and well as your experimental samples.
More Resources for Capturing the Best Publication-Quality Western Blot Images
- Handbook: Western Blotting Handbook
- Overview: Total Protein Normalization No-Stain Protein Labeling Reagent for Western Blotting
- Overview: Western Blot and Gel Imaging with iBright Imaging Systems
- App Note: Normalization of western blot data using No-Stain Protein Labeling Reagent and iBright 1500 Series Imaging Systems
- App Note: Getting publication-ready data from your western blotting experiments
- Peer-Reviewed Publication: “Biological validation of a novel process and product for quantitating western blots” (Journal of Biotechnology, 2021).
» Learn more about quantitative western blot imaging at thermofisher.com/ibright
Want to read more stories like this? Subscribe to Connect to Science, your portal for life science news.
##
References
“Artwork and Media Instructions | Elsevier Policy.” n.d. Beta.elsevier.com. Accessed August 24, 2023. https://beta.elsevier.com/about/policies-and-standards/author/artwork-and-media-instructions.
“Authors | Wiley.” n.d. Authorservices.wiley.com. Accessed August 24, 2023. https://authorservices.wiley.com/author-resources/index.html.
Begley, C. Glenn, and John P.A. Ioannidis. 2015. “Reproducibility in Science.” Circulation Research 116 (1): 116–26. https://doi.org/10.1161/circresaha.114.303819.
Carniol, Karen. 2015. “Common Pitfalls in Figure Preparation.” Cell Mentor. Cell Press and Cell Signaling Technology. April 15, 2015. https://crosstalk.cell.com/blog/common-pitfalls-in-figure-preparation.
“Cell Press: Cell Press.” n.d. Www.cell.com. Accessed August 24, 2023. https://www.cell.com/figureguidelines.
“Contributing to the Science Family of Journals.” n.d. Www.science.org. Accessed August 24, 2023. https://www.science.org/content/page/contributing-science-family-journals.
Diller, Thomas, Jordan Thompson, and Brian Steer. 2021. “Biological Validation of a Novel Process and Product for Quantitating Western Blots.” Journal of Biotechnology 326 (January): 52–60. https://doi.org/10.1016/j.jbiotec.2020.12.012.
Eaton, Samantha L., Sarah L. Roche, Maica Llavero Hurtado, Karla J. Oldknow, Colin Farquharson, Thomas H. Gillingwater, and Thomas M. Wishart. 2013. “Total Protein Analysis as a Reliable Loading Control for Quantitative Fluorescent Western Blotting.” Edited by Philipp J. Kahle. PLoS ONE 8 (8): e72457. https://doi.org/10.1371/journal.pone.0072457.
“Editorial Policies.” n.d. Www.asbmb.org. Accessed August 24, 2023. https://www.asbmb.org/journals-news/editorial-policies.
“Editorial Policies | Nature Portfolio.” n.d. Www.nature.com. Accessed August 24, 2023. https://www.nature.com/nature-portfolio/editorial-policies.
“Editorial Process.” 2022. American Association for Cancer Research Journals. 2022. Accessed August 24, 2023. https://aacrjournals.org/pages/editorial-process.
“For Authors | Nature.” n.d. Www.nature.com. Accessed August 24, 2023. https://www.nature.com/nature/for-authors.
Fosang, Amanda J., and Roger J. Colbran. 2015. “Transparency Is the Key to Quality.” Journal of Biological Chemistry 290 (50): 29692–94. https://doi.org/10.1074/jbc.E115.000002.
“Information for Authors .” n.d. Www.mdpi.com. Accessed August 24, 2023. https://www.mdpi.com/journal/biology/instructions.
“Information for Authors: Cell.” n.d. Www.cell.com. Accessed August 24, 2023. https://www.cell.com/cell/authors.
“Information for Authors: Journal of Biological Chemistry.” n.d. Www.jbc.org. Accessed August 24, 2023. https://www.jbc.org/content/authorinfo.
“Journal of Cell Biology Submission Guidelines.” 2023. Rockefeller University Press. 2023. Accessed August 24, 2023. https://rupress.org/jcb/pages/submission-guidelines.
Kroon, Cristina, Larissa Breuer, Lydia Jones, Jeehye An, Ayça Akan, Elkhansa Ahmed Mohamed Ali, Felix Busch, et al. 2022. “Blind Spots on Western Blots: Assessment of Common Problems in Western Blot Figures and Methods Reporting with Recommendations to Improve Them.” Edited by Mathieu J. M. Bertrand. PLOS Biology 20 (9): e3001783. https://doi.org/10.1371/journal.pbio.3001783.
“Promoting Integrity in Research and Its Publication | Committee on Publication Ethics: COPE.” 2019. Committee on Publication Ethics (COPE). 2019. Accessed August 24, 2023. https://publicationethics.org/.
© 2023 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified.
For Research Use Only. Not for use in diagnostic procedures.

Leave a Reply