 In a workshop held at the National Institutes of Health, representatives from across the globe met to discuss discrepancies in the ways targeted mass spectrometry (MS) assays are applied.1 Currently, no consensus exists on criteria to define the success of any one type of assay. In particular, some researchers have expressed concerns that targeted MS papers are published without clear and complete documentation of the analytical performance. Not only does this make it difficult to accurately review these papers for publication, but also, without a clear presentation of the methodology, researchers find it difficult to reproduce the data.
In a workshop held at the National Institutes of Health, representatives from across the globe met to discuss discrepancies in the ways targeted mass spectrometry (MS) assays are applied.1 Currently, no consensus exists on criteria to define the success of any one type of assay. In particular, some researchers have expressed concerns that targeted MS papers are published without clear and complete documentation of the analytical performance. Not only does this make it difficult to accurately review these papers for publication, but also, without a clear presentation of the methodology, researchers find it difficult to reproduce the data.
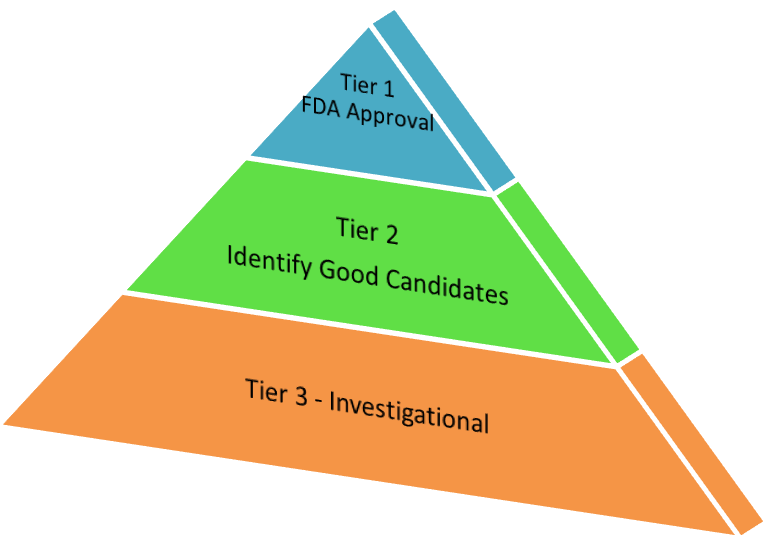
To remedy these concerns, the speakers and participants developed a “fit-for-purpose” approach identifying goals and characteristics of targeted MS assays. With an aim of improving these assays, they organized experimental design into three tiers.
Andy Hoofnagle (University of Washington) and Russell Grant (Laboratory Corporation of America) led the discussion of Tier 1 assays, with a goal of promoting accurate quantification. Two specific aims were (1) to provide accurate, precise, clinically actionable information for medical practitioners and (2) to inform decision making in the development of drugs for human use, such as for meeting the requirements of the U.S. Food and Drug Administration.
The scientists determined that the best practice in developing these assays was to use stable isotope-labeled internal standards for each target analyte or labeled analog proteins (if properly characterized and validated). They also recommended researchers apply multiple validation criteria—limit of detection (LOD), limit of the blank (LOB), lower limit of quantification (LLOQ) and calibration curves were among their ideas for building confidence and acceptability.
Representatives from the Broad Institute led the discussion of Tier 2 assays, whose goal is the accurate measurement of expression levels in proteins, peptides and protein modifications. Using target samples derived from cell lines, non-human animals or patient-derived materials, including tissues and biofluids, Tier 2 assays can be used to whittle down a long list of candidate biomarker proteins. The expression levels derived from discovery experiments can then help identify subsets of proteins truly reflecting disease presence, stage or response.
The speakers pointed out that in Tier 2 assays, the analytes are not well-characterized, unlike Tier 1 assays. Because of this, it is extremely important that the assays are highly sensitive, accurate and reproducible, so that researchers can be confident of their results.
The discussion leaders recommended isotope-labeled, internal standard peptides for each and every analyte. In addition, labeled concatamer peptides, “wing” peptides, other extended sequences or full-length proteins can improve precision.
Lastly, Bruno Domon (Luxembourg Clinical Proteomics Center) and Ruth Hüttenhain (UCSF, formerly ETH, Zurich) discussed Tier 3 measurements. What sets Tier 3 apart from the previously described Tier 1 and Tier 2 targeted assays is the absence of labeled internal standards for each analyte targeted. Without internal standards, researchers performing Tier 3 assays rely on chromatographic and mass spectrometric information to gain confidence in the methodology. Tier 3 studies are useful for early-stage biological studies and are not truly quantitative. Instead, they gain value through layers of evidence based on co-elution or overlap of fragment ions, the agreement of m/z and ratios of precursor or fragment ions, and reproducibility across replicates. Other clues can be gained through correlation of the observed retention with the predicted retention of known peptides.
Overall, the researchers call for all publications to include more detailed descriptions of the experimental methods used for analysis and quantification. As a result of these recommendations, Molecular and Cellular Proteomics plans to incorporate these guidelines for prospective submissions involving targeted MS studies.
Reference
1. Carr, S.A., et al. (2014, March) “Targeted peptide measurements in biology and medicine: Best practices for mass spectrometry-based assay development using a fit-for-purpose approach,” Molecular and Cellular Proteomics, 13(3) (pp. 907–17), doi: 10.1074/mcp.M113.036095.
Post Author: Emily Humphreys. Emily has previous research experience in eye development, infectious diseases, and aging. While she enjoyed the thrill of research, She has since traded bench work for science journalism. Emily has been a regular contributor to Accelerating Science since 2012.
Leave a Reply