Search Thermo Fisher Scientific

SuperScript IV Reverse Transcriptase |
SuperScript IV Reverse Transcriptase (RT) is a proprietary MMLV (Moloney Murine Leukemia Virus) RT mutant designed for reduced RNase H activity, increased thermostability, and highly efficient full-length cDNA synthesis. Compared to previous generation SuperScript RTs, SuperScript IV reverse transcriptase has significantly improved processivity, which enables fast reaction speed, inhibitor resistance, and exceptional performance even with challenging RNA samples. SuperScript IV reverse transcriptase is available in multiple formats tailored to specific applications, and it is widely cited in peer reviewed journals.
Advantages of SuperScript IV Reverse Transcriptase
SuperScript IV Reverse Transcriptase is known for its efficiency, sensitivity, robustness, short-reaction time, and thermostability. Click on the attributes below to see supporting data. For recommendations on choosing the reverse transcriptase fit for your application, use the selection tool.
- Efficient—Up to 100x higher cDNA yield
- Sensitive—Ct values reduced by as many as eight cycles for RT-qPCR
- Robust—Transcribes degraded and inhibitor-containing RNA samples
- Fast—10-minute cDNA synthesis time
- Stable—High thermostability to transcribe structured templates
SuperScript IV Reverse Transcriptase formats: Selection guide
SuperScript IV RT is available in several formats (Table 1)
- Stand-alone enzyme—Reverse transcriptase supplied with reaction buffer
- First-strand synthesis system—All cDNA synthesis reaction components are in separate tubes for maximum flexibility of reaction conditions
- Master mix—cDNA synthesis reaction components are premixed for exceptional efficiency and reduced variability in RT-qPCR applications
- One-step RT-PCR system—RT and PCR reagents for one-step RT-PCR applications
- Direct reverse transcription—Reagents for cell lysis and cDNA synthesis included; RNA purification is not required
Table 1. SuperScript IV RT formats.
| Product | Applications | Optimal reaction temperature | RT reaction time | Available formats |
|---|---|---|---|---|
| Most effective enzyme for all types of RNA, including difficult templates | 50–55°C | 10 min | Standalone enzyme Order now |
| First-strand cDNA synthesis reaction mix for two step RT-qPCR | 50°C | 10 min | |
| Flexibility in reaction conditions | 50–55°C | 10 min | ||
| Fast and simple RT-PCR workflow | 50–55°C | 10 min | SuperScript IV UniPrime One-step RT-PCR system (colored) SuperScript IV UniPrime One-step RT-PCR system (dye-free) |
SuperScript IV Single Cell/Low-Input cDNA PreAmp Kit
| cDNA synthesis and preamplification from single cells or low quantities of RNA | 50°C | 10 min | SuperScript IV Single Cell/Low-Input cDNA PreAmp Kit Order now |
| Direct cDNA synthesis from mammalian cells | 50°C | 10 min | SuperScript IV CellsDirect cDNA synthesis kit Order now |
| Full length cDNA synthesis with enhanced template switching efficiency | 30 min |
Comparison of SuperScript IV Reverse Transcriptase with other RTs
Stand-alone enzymes offer maximum flexibility in reaction setup. Table 2 compares the Superscript IV Reverse Transcriptase with other generations of reverse transcriptases.
Table 2. Comparison of standalone reverse transcriptases.
| M-MLV-Reverse Transcriptase | SuperScript II Reverse Transcriptase | Superscript III Reverse Transcriptase | Superscript IV Reverse Transcriptase | |
| Key attribute | Recombinant M-MLV RT for routine, non-demanding applications | Engineered M-MLV RT with reduced RNase H activity | Engineered M-MLV RT with reduced RNase H activity and improved thermal stability | Latest generation engineered RT for exceptional cDNA synthesis performance |
| Optimal reaction temperature | 37°C | 42°C | 50°C | 50–55°C |
| Reaction time | 50 min | 50 min | 30–50 min | 10 min |
| Sensitivity | 1 ng | 1 ng | 10 pg | 10 pg |
| Max cDNA length | Up to 7 kb | Up to 12.3 kb | Up to 12.3 kb | >12 kb |
| Ability to work with degraded or inhibitor-containing RNA | Low | Low | Medium | High |
| Reduced RNase H activity | No | Yes | Yes | Yes |
Performance of SuperScript IV Reverse Transcriptase
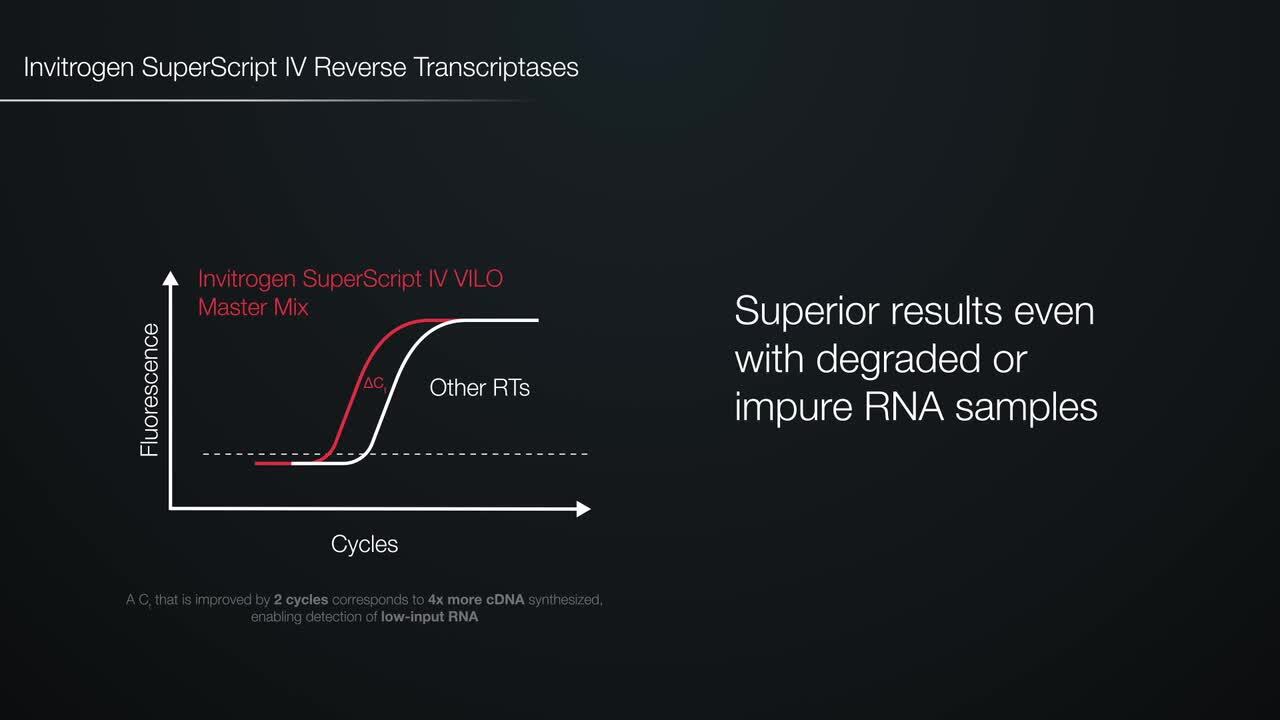
Up to 100x higher cDNA yields with degraded RNA (Figure 1).
Figure 1. High efficiency with degraded RNA. RT-qPCR of degraded RNA (RIN 1–3) from human cells and plant tissues was performed with different brands of commercially available RTs and Applied Biosystems TaqMan assays. Delta Ct values (ΔCt = Ct – Ct SuperScript IV) show that SuperScript IV RT delivered up to 100x higher cDNA yields and has lower Ct values compared to SuperScript III and other commercial reverse transcriptases.
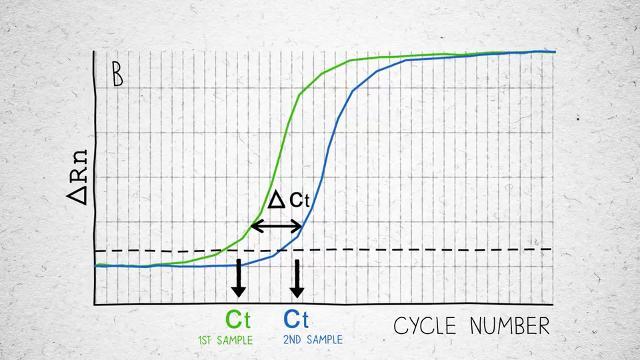
Low Ct values. Ct values reduced by 8 cycles compared to other reverse transcriptases (Figure 2).
Figure 2. Sensitivity and variability in cDNA synthesis using degraded RNA samples. Degraded Arabidopsis total RNA (RIN: 1–3), in amounts of 1, 10, and 100 ng were used as input RNA in 20 μL reverse transcription reactions with SuperScript IV Reverse Transcriptase and random hexamers according to the product protocol. RTs from other vendors were used according to the manufacturers’ protocols. For each tested enzyme and each RNA input, RT reactions were performed in triplicates. From each reverse transcription reaction, 10% of the cDNA product was added to TaqMan assays for two targets, Gln synthetase and WRKY TF 70. Three qPCR reactions were performed for each reverse transcription reaction and the average Ct values for each RNA input were plotted (standard deviation from 9 Ct values for each input RNA).
Compounds that have inhibitory effects on RTs are commonly found in RNA samples even after thorough purification. These compounds may interfere with cDNA synthesis, produce false RT-PCR and RT-qPCR results, and cause results to be misinterpreted. RT inhibitors include reagents used during RNA extraction, and co-purified contaminants arising from biological samples (Table 3). SuperScript IV Reverse Transcriptase shows significantly improved resistance to contaminating inhibitors when compared to previous generation SuperScript III Reverse Transcriptase and other commercially available RTs (Figure 3).
Table 3. Common cDNA synthesis inhibitors and their sources.
| Inhibitor | Source |
|---|---|
| Ethanol/isopropanol, salts, phenol/chloroform, detergents | Sample preparation |
| Hematin, bile salts | Blood, feces |
| Humic acid, polyphenols, polysaccharides | Soil, plants |
| Formalin, paraffin | FFPE |
Figure 3. Higher performance in cDNA synthesis in the presence of biological or sample prep inhibitors. A 0.5–10 kb RNA ladder was used in a 10 μL SuperScript IV Reverse Transcriptase reaction with oligo(dT)20 according to the product protocol. RTs from other vendors were used according to their respective protocols. Inhibitors were added to the RNA samples prior to primer annealing or addition of RT reaction mix. First-strand cDNAs were resolved by alkaline gel electrophoresis, and cDNA was stained using Invitrogen SYBR Gold Nucleic Acid Gel Stain. During electrophoresis, NaOH hydrolyzes all RNA, resulting in visualization of cDNA only.
SuperScript IV Reverse Transcriptase has remarkable processivity and thereby generates long, full-length cDNA fragments within a short reaction time. Figure 4 demonstrates that SuperScript IV Reverse Transcriptase synthesized cDNAs of up to 9 kb in 10 minutes, while most other commercially available RTs were only able to synthesize cDNAs between 1.5–3 kb or less in the same duration.
Figure 4. Fast cDNA synthesis capability. The Invitrogen Millennium RNA Marker was used in a 10 μL reaction with SuperScript IV Reverse Transcriptase and oligo(dT) primer according to the product protocol. Other commercially available RTs were used according to manufacturers’ protocols, except for reaction times reduced to 10 minutes. First-strand cDNAs were resolved by alkaline gel electrophoresis, and cDNA was stained using SYBR Gold Nucleic Acid Gel Stain. During electrophoresis NaOH hydrolyzes all RNA, resulting in visualization of cDNA only.
When reverse transcription reactions are performed at low temperatures (less than 42°C), RNA secondary structures, especially GC-rich templates, may interfere with cDNA synthesis. SuperScript IV Reverse Transcriptase has high thermostability and can be used in reactions at 50°C or higher, which facilitates the successful transcription of highly structured RNA transcripts (Figure 5).
Figure 5. High thermostability of SuperScript IV Reverse Transcriptase. A 0.5–10 kb RNA ladder was used in a 10 μL SuperScript IV RT reaction with oligo(dT) according to the product protocol, with the exception that reaction temperature was varied between 50 and 65°C. First-strand cDNAs were resolved by alkaline gel electrophoresis, and cDNA was stained using SYBR Gold Nucleic Acid Gel Stain. During electrophoresis NaOH hydrolyzes all RNA, resulting in the visualization of cDNA only. cDNA bands were quantitated by TotalLab software for each reaction temperature. Percentage SuperScript IV RT activity was calculated by dividing values at each reaction temperature by values at 50°C.
Up to 100x higher cDNA yields with degraded RNA (Figure 1).
Figure 1. High efficiency with degraded RNA. RT-qPCR of degraded RNA (RIN 1–3) from human cells and plant tissues was performed with different brands of commercially available RTs and Applied Biosystems TaqMan assays. Delta Ct values (ΔCt = Ct – Ct SuperScript IV) show that SuperScript IV RT delivered up to 100x higher cDNA yields and has lower Ct values compared to SuperScript III and other commercial reverse transcriptases.
Low Ct values. Ct values reduced by 8 cycles compared to other reverse transcriptases (Figure 2).
Figure 2. Sensitivity and variability in cDNA synthesis using degraded RNA samples. Degraded Arabidopsis total RNA (RIN: 1–3), in amounts of 1, 10, and 100 ng were used as input RNA in 20 μL reverse transcription reactions with SuperScript IV Reverse Transcriptase and random hexamers according to the product protocol. RTs from other vendors were used according to the manufacturers’ protocols. For each tested enzyme and each RNA input, RT reactions were performed in triplicates. From each reverse transcription reaction, 10% of the cDNA product was added to TaqMan assays for two targets, Gln synthetase and WRKY TF 70. Three qPCR reactions were performed for each reverse transcription reaction and the average Ct values for each RNA input were plotted (standard deviation from 9 Ct values for each input RNA).
Compounds that have inhibitory effects on RTs are commonly found in RNA samples even after thorough purification. These compounds may interfere with cDNA synthesis, produce false RT-PCR and RT-qPCR results, and cause results to be misinterpreted. RT inhibitors include reagents used during RNA extraction, and co-purified contaminants arising from biological samples (Table 3). SuperScript IV Reverse Transcriptase shows significantly improved resistance to contaminating inhibitors when compared to previous generation SuperScript III Reverse Transcriptase and other commercially available RTs (Figure 3).
Table 3. Common cDNA synthesis inhibitors and their sources.
| Inhibitor | Source |
|---|---|
| Ethanol/isopropanol, salts, phenol/chloroform, detergents | Sample preparation |
| Hematin, bile salts | Blood, feces |
| Humic acid, polyphenols, polysaccharides | Soil, plants |
| Formalin, paraffin | FFPE |
Figure 3. Higher performance in cDNA synthesis in the presence of biological or sample prep inhibitors. A 0.5–10 kb RNA ladder was used in a 10 μL SuperScript IV Reverse Transcriptase reaction with oligo(dT)20 according to the product protocol. RTs from other vendors were used according to their respective protocols. Inhibitors were added to the RNA samples prior to primer annealing or addition of RT reaction mix. First-strand cDNAs were resolved by alkaline gel electrophoresis, and cDNA was stained using Invitrogen SYBR Gold Nucleic Acid Gel Stain. During electrophoresis, NaOH hydrolyzes all RNA, resulting in visualization of cDNA only.
SuperScript IV Reverse Transcriptase has remarkable processivity and thereby generates long, full-length cDNA fragments within a short reaction time. Figure 4 demonstrates that SuperScript IV Reverse Transcriptase synthesized cDNAs of up to 9 kb in 10 minutes, while most other commercially available RTs were only able to synthesize cDNAs between 1.5–3 kb or less in the same duration.
Figure 4. Fast cDNA synthesis capability. The Invitrogen Millennium RNA Marker was used in a 10 μL reaction with SuperScript IV Reverse Transcriptase and oligo(dT) primer according to the product protocol. Other commercially available RTs were used according to manufacturers’ protocols, except for reaction times reduced to 10 minutes. First-strand cDNAs were resolved by alkaline gel electrophoresis, and cDNA was stained using SYBR Gold Nucleic Acid Gel Stain. During electrophoresis NaOH hydrolyzes all RNA, resulting in visualization of cDNA only.
When reverse transcription reactions are performed at low temperatures (less than 42°C), RNA secondary structures, especially GC-rich templates, may interfere with cDNA synthesis. SuperScript IV Reverse Transcriptase has high thermostability and can be used in reactions at 50°C or higher, which facilitates the successful transcription of highly structured RNA transcripts (Figure 5).
Figure 5. High thermostability of SuperScript IV Reverse Transcriptase. A 0.5–10 kb RNA ladder was used in a 10 μL SuperScript IV RT reaction with oligo(dT) according to the product protocol, with the exception that reaction temperature was varied between 50 and 65°C. First-strand cDNAs were resolved by alkaline gel electrophoresis, and cDNA was stained using SYBR Gold Nucleic Acid Gel Stain. During electrophoresis NaOH hydrolyzes all RNA, resulting in the visualization of cDNA only. cDNA bands were quantitated by TotalLab software for each reaction temperature. Percentage SuperScript IV RT activity was calculated by dividing values at each reaction temperature by values at 50°C.
Resources for SuperScript IV Reverse Transcriptase
Interactive selection tool: Find the right reverse transcriptase for your experiments.
Watch this video on the advantages of SuperScript IV Reverse Transcriptase.
Understand how SuperScript IV Reverse Transcriptase consistently delivers low Ct values in qPCR reactions.
Discover how SuperScript IV Reverse Transcriptase performs despite presence of inhibitors.
Learn how the SuperScript IV UniPrime One-Step RT-PCR System makes it easy to set up reactions while bringing superior results.
Manuals
Protocols
Technical articles
SuperScript IV Reverse Transcriptase FAQs
Find tips, troubleshooting help, and resources for common questions about SuperScript Reverse Transcriptases. Can’t find your question? Search the FAQ database
The SuperScript IV enzyme has been engineered for higher thermostability, processivity, and cDNA yields. It performs better in the presence of inhibitors, and the reaction buffer has also been optimized for robust cDNA synthesis from a wide range of samples.
When compared with SuperScript III RT (and other manufacturers’ RTs) in a synthesis reaction for a 9 kb cDNA, SuperScript IV RT performed successful synthesis in just 10 minutes and did so with comparable (or improved) yield.
SuperScript IV RT sustains 100% activity at up to 56.4°C and 70% activity at up to 65°C, while wild type MMLV RT or MMLV RNase H– RT enzymes usually display very low or no activity above 45°C. SuperScript IV RT’s ability to function at higher temperatures enables the reverse transcription of RNA targets with structural complexities.
SuperScript IV Reverse Transcriptases: Citations
SuperScript IV reverse transcriptases are highly cited in the several peer reviewed research publications. In three years, between 2022 and 2024, it has been cited over in 10K publications.
| Use | Reference |
|---|---|
Detect cancer biomarkers from extracellular vesicles by ddPCR (digital droplet). | Lee KY, Beatson EL, Knechel MA et al. (2024). Detection of Extracellular Vesicle-Derived RNA as Potential Prostate Cancer Biomarkers: Role of Cancer-type SLCO1B3 and ABCC3. J Cancer 15(3):615–622. doi: 10.7150/jca.90836. PMID: 38213719. |
cDNA synthesis for preparation of a Poly(A)-seq library and also for nested RT-PCR to study changes in UTR length. | Gabel AM, Belleville AE, Thomas JD et al. (2024). Multiplexed screening reveals how cancer-specific alternative polyadenylation shapes tumor growth in vivo.Nat Commun 15:959. doi: 10.1038/s41467-024-44931-x. PMID: 38302465 |
Synthesize cDNA from FFPE samples for ddPCR. | Look T, Puca E, Bühler M et al. (2023). Targeted delivery of tumor necrosis factor in combination with CCNU induces a T cell-dependent regression of glioblastoma. Sci Transl Med 15(697). doi:10.1126/scitranslmed.adf2281 PMID: 37224228 |
| Use | Reference |
|---|---|
Study mRNA expression profiles in tissue and identify a unique pathway in a shark species. | Cutler CP, Omoregie E, Ojo T. UT-1 Transporter Expression in the Spiny Dogfish (Squalus acanthias): UT-1 Protein Shows a Different Localization in Comparison to That of Other Sharks. Biomolecules 14(9):1151. doi: 10.3390/biom14091151. PMID: 39334917 |
Used SuperScript IV Reverse Transcriptase with low input sample difficult sample (RNA from insect antennae). | Johny J, Nihad M, Alharbi HA et al. (2024). Silencing sensory neuron membrane protein RferSNMPu1 impairs pheromone detection in the invasive Asian Palm Weevil.Sci Rep 14(1):16541. doi: 10.1038/s41598-024-67309-x. PMID: 39019908 |
Synthesize cDNA from RNA isolated from yeast cells to measure gene expression. | Montrose K, Lac DT, Burnetti AJ et al.(2024). Proteostatic tuning underpins the evolution of novel multicellular traits. Sci Adv 10(10). doi:10.1126/sciadv.adn2706 PMID: 38457507 |
| Use | Reference |
|---|---|
Convert RNA to cDNA prior to a qPCR reaction to measure the relative expression levels of target genes in wound healing assays. | Rapp J, Ness J, Wolf J (2024). 2D and 3D in vitro angiogenesis assays highlight different aspects of angiogenesis. Biochim Biophys Acta Mol Basis Dis 1870(3). doi: 10.1016/j.bbadis PMID: 38244944 |
Synthesize cDNA for the study of RNA splicing. Products were analyzed by agarose gel electrophoresis, qPCR, and Sanger sequencing. | Žedaveinytė R, Meers C, Le HC et al. (2024). Antagonistic conflict between transposon-encoded introns and guide RNAs. Science 385(6705). doi:10.1126/science.adm8189 PMID: 38991068 |
Synthesize cDNA from CRISPR modifed cells for RNA-seq. | Dudnyk K, Cai D, Shi C, Xu J et al. (2024). Sequence basis of transcription initiation in the human genome. Science 384(6694). doi:10.1126/science.adj0116 PMID: 38662817 |
| Use | Reference |
|---|---|
Detect cancer biomarkers from extracellular vesicles by ddPCR (digital droplet). | Lee KY, Beatson EL, Knechel MA et al. (2024). Detection of Extracellular Vesicle-Derived RNA as Potential Prostate Cancer Biomarkers: Role of Cancer-type SLCO1B3 and ABCC3. J Cancer 15(3):615–622. doi: 10.7150/jca.90836. PMID: 38213719. |
cDNA synthesis for preparation of a Poly(A)-seq library and also for nested RT-PCR to study changes in UTR length. | Gabel AM, Belleville AE, Thomas JD et al. (2024). Multiplexed screening reveals how cancer-specific alternative polyadenylation shapes tumor growth in vivo.Nat Commun 15:959. doi: 10.1038/s41467-024-44931-x. PMID: 38302465 |
Synthesize cDNA from FFPE samples for ddPCR. | Look T, Puca E, Bühler M et al. (2023). Targeted delivery of tumor necrosis factor in combination with CCNU induces a T cell-dependent regression of glioblastoma. Sci Transl Med 15(697). doi:10.1126/scitranslmed.adf2281 PMID: 37224228 |
| Use | Reference |
|---|---|
Study mRNA expression profiles in tissue and identify a unique pathway in a shark species. | Cutler CP, Omoregie E, Ojo T. UT-1 Transporter Expression in the Spiny Dogfish (Squalus acanthias): UT-1 Protein Shows a Different Localization in Comparison to That of Other Sharks. Biomolecules 14(9):1151. doi: 10.3390/biom14091151. PMID: 39334917 |
Used SuperScript IV Reverse Transcriptase with low input sample difficult sample (RNA from insect antennae). | Johny J, Nihad M, Alharbi HA et al. (2024). Silencing sensory neuron membrane protein RferSNMPu1 impairs pheromone detection in the invasive Asian Palm Weevil.Sci Rep 14(1):16541. doi: 10.1038/s41598-024-67309-x. PMID: 39019908 |
Synthesize cDNA from RNA isolated from yeast cells to measure gene expression. | Montrose K, Lac DT, Burnetti AJ et al.(2024). Proteostatic tuning underpins the evolution of novel multicellular traits. Sci Adv 10(10). doi:10.1126/sciadv.adn2706 PMID: 38457507 |
| Use | Reference |
|---|---|
Convert RNA to cDNA prior to a qPCR reaction to measure the relative expression levels of target genes in wound healing assays. | Rapp J, Ness J, Wolf J (2024). 2D and 3D in vitro angiogenesis assays highlight different aspects of angiogenesis. Biochim Biophys Acta Mol Basis Dis 1870(3). doi: 10.1016/j.bbadis PMID: 38244944 |
Synthesize cDNA for the study of RNA splicing. Products were analyzed by agarose gel electrophoresis, qPCR, and Sanger sequencing. | Žedaveinytė R, Meers C, Le HC et al. (2024). Antagonistic conflict between transposon-encoded introns and guide RNAs. Science 385(6705). doi:10.1126/science.adm8189 PMID: 38991068 |
Synthesize cDNA from CRISPR modifed cells for RNA-seq. | Dudnyk K, Cai D, Shi C, Xu J et al. (2024). Sequence basis of transcription initiation in the human genome. Science 384(6694). doi:10.1126/science.adj0116 PMID: 38662817 |
Resources
Reverse transcriptase selection tool
Find the right products to fit your cDNA synthesis needs.
Reverse transcriptase webinars
Stay up-to-date on current industry trends in cDNA synthesis applications with on-demand webinars.
Reverse transcriptase literature
Access white papers and technical and application notes for insights on cutting edge reverse transcription technology.
Reverse transcriptase videos
Watch instructional videos on troubleshooting tips, how-to videos, and explainers related to cDNA synthesis.
Reverse transcriptase education
Review educational resources on topics including reverse transcription basics, cDNA synthesis, enzyme selection, troubleshooting tips, and cDNA applications in molecular biology.
Support
PCR and cDNA Synthesis Support Center
Find support for every step of your cDNA synthesis experiments.
Tm calculator
Handy tool to calculate Tm (melting temperature) of primers and annealing temperatures.
Contact us
Get in touch with our technical application scientists for questions regarding reverse transcription enzymes, master mixes, and cDNA synthesis kits.
Mobile and desktop applications
Access content and helpful tools through SmartIST and Cloning Bench mobile apps.
OEM and custom solutions
Source reverse transcriptase and other molecular biology enzymes, PCR reagents, and plastics customized to your needs.
For Research Use Only. Not for use in diagnostic procedures.